
Our Technology
Ion channels play a fundamental role in physiological functions. They are also extremely important drug targets because of their role in numerous neurological, cardiovascular, and metabolic diseases, cancer cell proliferation, metastasis, and immune cell regulation.
From the 1950s to the 1980s, many drugs targeting ion channels were developed, still in use today. However, most of them lack good selectivity for their target molecules. Thus, their use and side effects must be carefully controlled. Given these problems, developing ion channel drugs with high target selectivity is a critical topic in the pharmaceutical industry, and much effort has been devoted to addressing this issue. However, even in recent years, many ion channel drugs have been developed as me-too drugs with only minor modifications in function and properties based on the original core structure. Unfortunately, there have been only a few successful ion channel drugs with novel structures and excellent activity and selectivity.
Let’s look at some of the creatures in nature. Species such as spiders, scorpions, snakes, and cone shells have survived in the fierce survival race by harnessing their venom. These species prey on insects and animals by injecting venom into their prey, paralyzing their nerves and making them unable to move. The venom contains a variety of peptides and proteins, each with highly bioactive properties. Interestingly, many of the peptides in venom have potent activity and relatively high selectivity, especially those that act on ion channels. They are called disulfide-rich peptides (DRPs) because their molecules have multiple disulfide bonds. In recent years, many DRPs have been discovered in the venom of various organisms, and many researchers have shown that each has a unique amino acid sequence and functions as an ion channel modulator. For example, GTx1-15, found in the venom of the rose hair tarantula, selectively inhibits the function of the voltage-gated sodium channels Nav1.7 and Nav1.3, which are considered promising target molecules for the development of analgesics. ShK, found in the venom of the sea anemone Stichodactyla helianthus, is a potent inhibitor of the function of voltage-gated potassium channels (Kv1.1, Kv1.3, Kv1.4, and Kv1.6).
These natural DRPs are considered promising lead compounds for ion channel drug development because they regulate specific ion channel functions with high potency and relatively high selectivity. Based on this idea, an analgesic called ziconotide has been developed from the cone shell-derived DRP ω-conotoxin. Ziconotide is a potent inhibitor of neuronal N-type voltage-sensitive calcium channels and is effective against severe pain that cannot be treated with morphine.
Ziconotide is the only ion channel drug developed based on DRPs. Unfortunately, despite great promise, most DRP drug development efforts have been unsuccessful. One reason is that DRPs have complex structures and are extremely difficult to synthesize chemically. Because of the difficulty of synthesis, the number of amino acid substitutions that can be evaluated for activity and selectivity is extremely limited, making it challenging to design DRPs with sufficient activity and selectivity for drug development.
To overcome these difficulties in DRP drug development, we have developed the Veneno Suite, an innovative DRP screening technology that is distinctly different from conventional DRP discovery methods. We will leverage this Veneno Suite to develop ion channel drugs with high potency and selectivity, which were impossible to develop with small molecule compounds or antibodies.
The Veneno Suite also applies to drug discovery targeting membrane proteins such as GPCRs and transporters. We will deploy drug discovery for various diseases by targeting membrane proteins, which has been challenging for drug discovery.
Furthermore, we leverage genomic information to discover functional DRPs targeting membrane proteins in animals, insects, plants, and bacteria. Through collaboration with various companies, we will also work on developing veterinary drugs, agrochemical ingredients, and functional biochemical materials.
Veneno Suite
Natural DRPs have evolved to be potent against animal and insect ion channels but have not been optimized as modulators for human ion channels; to make DRPs into drugs, they must be engineered.
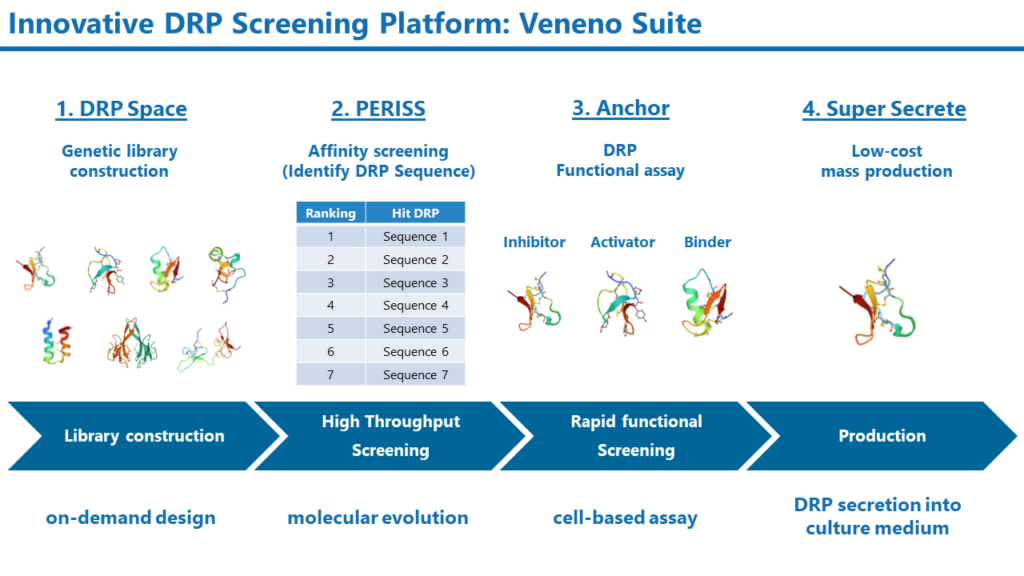
1. DRP Space:
Method for constructing a genetic DRP library with enormous diversity.
2. PERISS:
High-throughput screening method for DRPs that employs evolutionary molecular engineering
3. Anchor:
Cell-based assay for rapid evaluation of DRP activity and selectivity
4. Super Secrete:
Low-cost production of DRPs using E. coli
The Veneno Suite enables the construction of DRP genetic libraries based on DRP genomic, structural, and physicochemical information. The DRP Space provides libraries of enormous diversity, which conventional chemical synthesis methods cannot construct. The constructed genetic libraries are then subjected to PERISS, a high-throughput screening method that leverages evolutionary molecular engineering. DRPs with high affinity to the target membrane protein will be selected from a billion-diverse DRP library in PERISS.
Their activity will be rapidly screened by the Anchor method. The Anchor uses assay methods specific to the target molecule. For example, when targeting ion channels, the function of DRPs is identified by electrophysiological methods.
Super Secrete is an E. coli-based method for DRP production that allows DRPs to be folded in periplasmic space and secreted into the culture medium. This method enables DRPs to be produced more rapidly and inexpensively than conventional chemical synthesis methods.
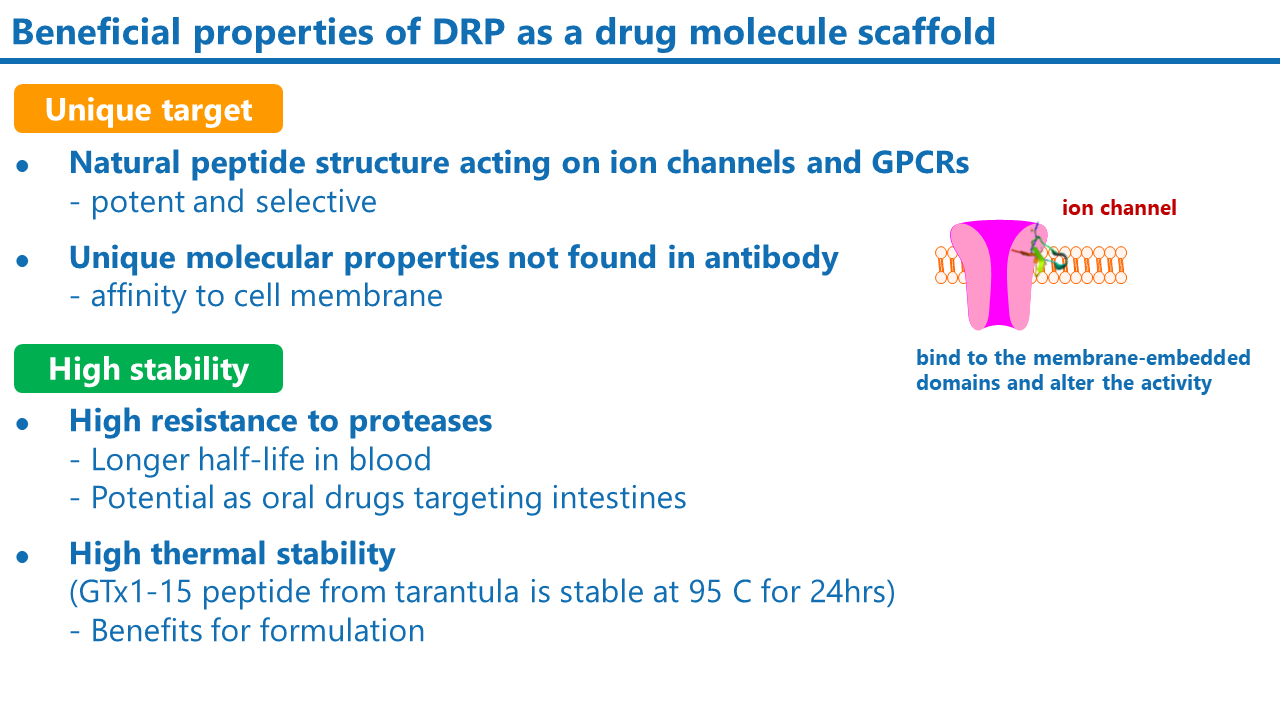
What is Disulfide-Rich Peptide (DRP)?
Disulfide-rich peptides (DRPs) have multiple disulfide bonds in the molecule. They are usually composed of 20 to 60 amino acids. DRPs, especially those found in the venom of spiders, scorpions, and snakes, are known to be potent and selective against ion channels and GPCRs. The compact and stable structure of DRPs also makes them highly stable against digestive enzymes, acids, and heat, compared to linear peptides. DRPs are also known to be less immunogenic due to their stability. These properties make DRP a promising lead compound for ion channel and GPCR drug development.
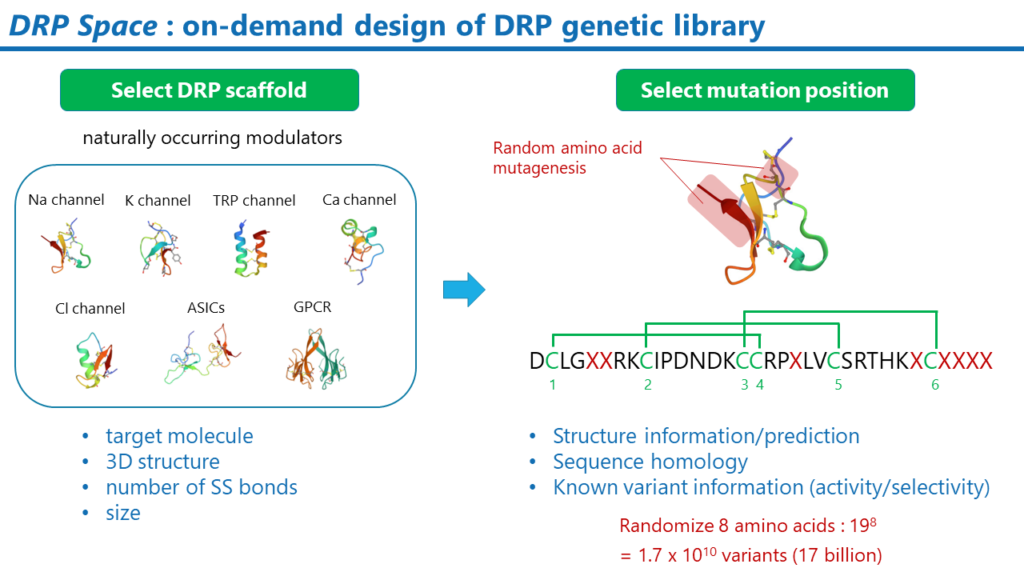
DRPSpace
DRP Space is a method of constructing DRP genetic libraries which will be designed for each drug target.
DRP Libraries can be constructed based on the drug target or drug design concept. For example, if sodium channels are the drug target, the natural sodium channel modulator DRP could be a good candidate for the scaffold.
Once the DRP scaffold is selected, the positions where random amino acid mutations are to be introduced are determined. The position of the mutation can be determined based on different ideas. One idea is to select the potential sites for interaction with a membrane protein. The potential interaction sites will be estimated using computational methods such as the Membrane Docking Optimal Area method. Public information on the activity and selectivity of known DRP mutants can also be helpful.
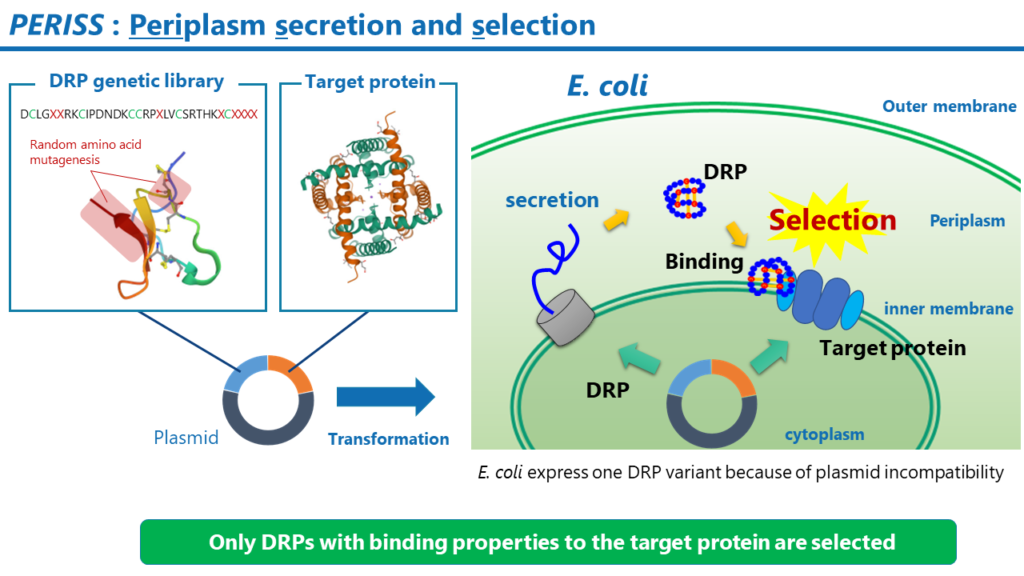
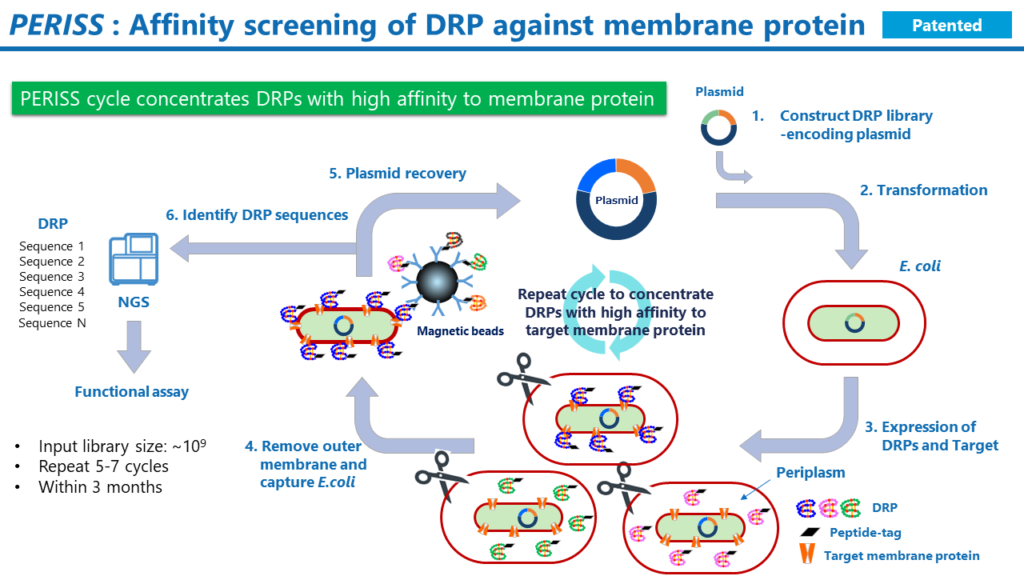
PERISS
PERISS is a high-throughput screening method based on evolutionary molecular engineering. The PERISS method enriches plasmids encoding DRPs that bind strongly to target membrane proteins. By analyzing the DNA sequence encoding the DRP in the enriched plasmid using a next-generation sequencer, the amino acid sequence of the DRP that can bind strongly to the target membrane protein is identified.
The process is as follows.
1. Prepare plasmids encoding the DRP genetic library and the target protein.
2. Transform E. coli with the prepared plasmid.
3. Culture the transformed E. coli. The target protein expresses on the inner membrane of the E. coli, and the DRP expresses in the periplasmic space; the disulfide bonds in the DRP are formed by the Dsb family, a group of enzymes that promote disulfide bond formation. Due to plasmid incompatibility, only one variant of DRP expresses in a single E. coli.
4. Remove the outer membrane of the E. coli and collect the E. coli spheroplasts with magnetic beads labeled with an antibody against the tag sequence added to the end of the DRP. This procedure recovers E. coli that carries a plasmid encoding a DRP that binds to the target membrane protein.
5. Extract the plasmid from the E. coli and transform the E. coli again with the resulting plasmid.
6. The above procedure is repeated 5 to 7 times to enrich plasmids encoding a DRP that binds strongly to the target membrane protein. Finally, the DNA sequences encoding the DRP in the recovered plasmids are analyzed on a next-generation sequencer to identify the amino acid sequences. Then, in the next step, identify the function of DRPs in the sequences with a higher frequency of occurrence among the NGS reads.
One billion sequences are input to the PERISS screening from the DRP genetic library constructed by DRP Space. Due to plasmid incompatibility, the PERISS method uses one E. coli as a reaction field to select DRP mutants with high affinity to the target membrane protein. The entire process can be completed in three months.
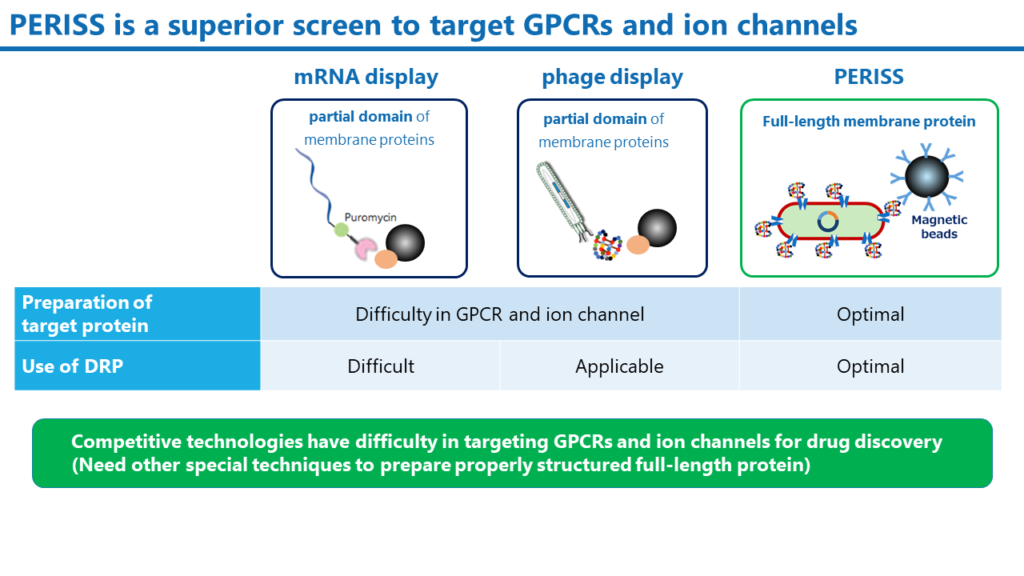
The PERISS method is a unique technology that uses the periplasmic space of E. coli, where DRP molecules translated in the cytoplasm correctly construct disulfide bonds. Full-length target membrane proteins can be expressed on the inner membrane of E. coli. On the other hand, mRNA display and phage display, which are widely used antibody and peptide screening methods, require different and very sophisticated techniques to express full-length membrane proteins on lipid bilayers. In addition, mRNA display is unsuitable for screening DRP molecules because of the difficulty in forming disulfide bonds for DRPs.
The PERISS method is not technology-specific only to ion channels drug discovery. We have already demonstrated the ability to express various human membrane proteins, including GPCRs, and even soluble proteins can be targeted by anchoring them on the inner membrane. The PERISS method is a versatile screening method that can identify DRPs that bind to various proteins.
PERISS
PERISS is a high-throughput screening method based on evolutionary molecular engineering. The PERISS method enriches plasmids encoding DRPs that bind strongly to target membrane proteins. By analyzing the DNA sequence encoding the DRP in the enriched plasmid using a next-generation sequencer, the amino acid sequence of the DRP that can bind strongly to the target membrane protein is identified.
The process is as follows.
1. Prepare plasmids encoding the DRP genetic library and the target protein.
2. Transform E. coli with the prepared plasmid.
3. Culture the transformed E. coli. The target protein expresses on the inner membrane of the E. coli, and the DRP expresses in the periplasmic space; the disulfide bonds in the DRP are formed by the Dsb family, a group of enzymes that promote disulfide bond formation. Due to plasmid incompatibility, only one variant of DRP expresses in a single E. coli.
4. Remove the outer membrane of the E. coli and collect the E. coli spheroplasts with magnetic beads labeled with an antibody against the tag sequence added to the end of the DRP. This procedure recovers E. coli that carries a plasmid encoding a DRP that binds to the target membrane protein.
5. Extract the plasmid from the E. coli and transform the E. coli again with the resulting plasmid.
6. The above procedure is repeated 5 to 7 times to enrich plasmids encoding a DRP that binds strongly to the target membrane protein. Finally, the DNA sequences encoding the DRP in the recovered plasmids are analyzed on a next-generation sequencer to identify the amino acid sequences. Then, in the next step, identify the function of DRPs in the sequences with a higher frequency of occurrence among the NGS reads.
One billion sequences are input to the PERISS screening from the DRP genetic library constructed by DRP Space. Due to plasmid incompatibility, the PERISS method uses one E. coli as a reaction field to select DRP mutants with high affinity to the target membrane protein. The entire process can be completed in three months.
The PERISS method is a unique technology that uses the periplasmic space of E. coli, where DRP molecules translated in the cytoplasm correctly construct disulfide bonds. Full-length target membrane proteins can be expressed on the inner membrane of E. coli. On the other hand, mRNA display and phage display, which are widely used antibody and peptide screening methods, require different and very sophisticated techniques to express full-length membrane proteins on lipid bilayers. In addition, mRNA display is unsuitable for screening DRP molecules because of the difficulty in forming disulfide bonds for DRPs.
The PERISS method is not technology-specific only to ion channels drug discovery. We have already demonstrated the ability to express various human membrane proteins, including GPCRs, and even soluble proteins can be targeted by anchoring them on the inner membrane. The PERISS method is a versatile screening method that can identify DRPs that bind to various proteins.



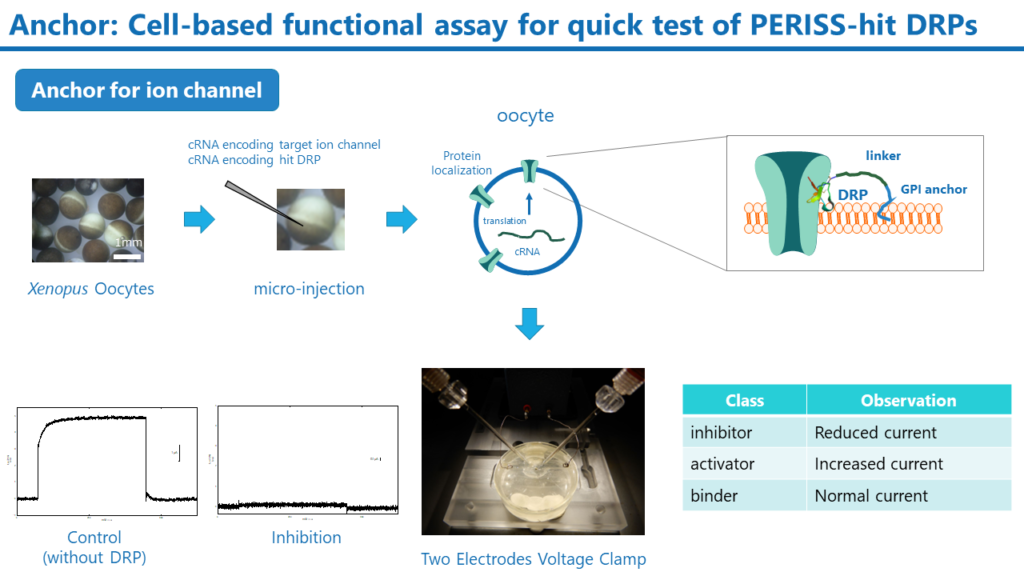
Anchor
The PERISS method identifies many different DRP sequences with binding properties to target membrane proteins. Anchor is a rapid assay to determine whether these hit DRPs can regulate the target membrane protein function. The Anchor requires the preparation of cRNAs for drug target membrane proteins and hit DRPs. Both cRNAs are then microinjected into the Xenopus Oocytes, and the membrane protein and DRP are expressed on the cell membrane of the oocytes. If the target membrane protein is ion channel, the DRP function can be evaluated by measuring ion channel activity using the two-electrode voltage clamp method.
Although the Anchor is a semi-quantitative method, it can rapidly evaluate the activity of DRPs by simply preparing the cRNA of DRP without preparing individual DRP peptides. In addition, target selectivity can also be evaluated by expressing ion channels other than the target. DRPs with high activity are produced by the Super Secrete method described below, and their IC50 and EC50 are analyzed in detail using cell and animal-based assays.
Anchor
The PERISS method identifies many different DRP sequences with binding properties to target membrane proteins. Anchor is a rapid assay to determine whether these hit DRPs can regulate the target membrane protein function. The Anchor requires the preparation of cRNAs for drug target membrane proteins and hit DRPs. Both cRNAs are then microinjected into the Xenopus Oocytes, and the membrane protein and DRP are expressed on the cell membrane of the oocytes. If the target membrane protein is ion channel, the DRP function can be evaluated by measuring ion channel activity using the two-electrode voltage clamp method.
Although the Anchor is a semi-quantitative method, it can rapidly evaluate the activity of DRPs by simply preparing the cRNA of DRP without preparing individual DRP peptides. In addition, target selectivity can also be evaluated by expressing ion channels other than the target. DRPs with high activity are produced by the Super Secrete method described below, and their IC50 and EC50 are analyzed in detail using cell and animal-based assays.

Super Secrete
DRPs have been produced mainly by chemical synthesis (peptide synthesis). However, the chemical synthesis method makes it difficult to synthesize large amounts of DRPs with multiple disulfide bonds correctly formed in the molecule and requires high production costs. This has been a significant hurdle for DRP drug discovery in speeding up pharmacological evaluation and toxicity/safety assessment.
In collaboration with a Japanese chemical company, we have developed the Super Secrete method, a technology to express DRPs in the periplasmic space of E. coli and secrete them into the culture medium after folding them into the correct structure.
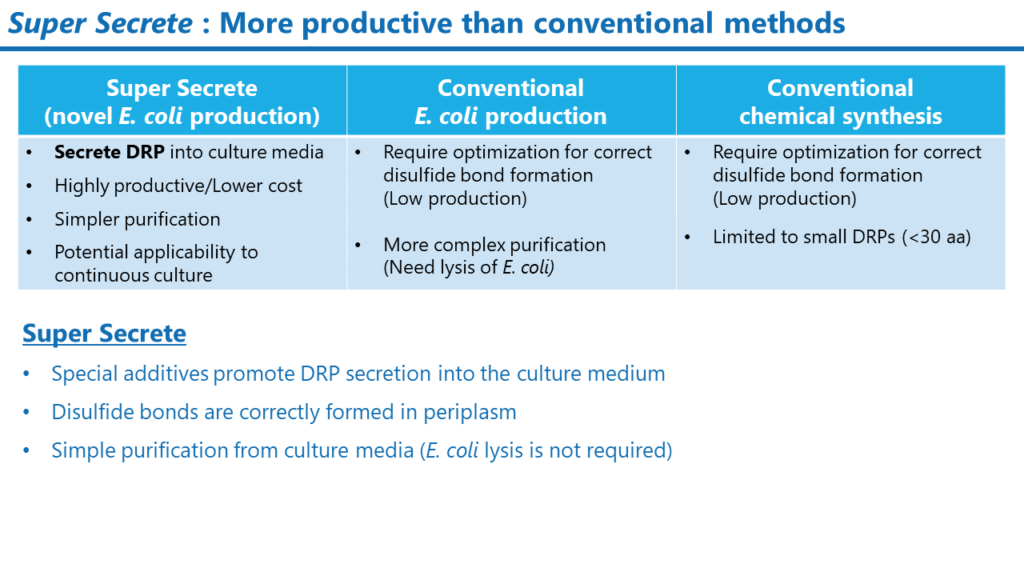
Generally, E. coli cannot secrete peptides or proteins into the culture medium, but the Super Secrete Method makes this possible using a unique culture medium. In purifying peptides and proteins by disrupting E. coli, a careful purification process is required to avoid contamination of E. coli-derived components such as LPS. Super Secrete, on the other hand, does not require disruption of E. coli, thus significantly reducing contamination of bacteria-derived components and simplifying the purification process. At the laboratory level, we have established a small-scale production method (culture and purification using a small jar fermenter).
Super Secrete enables us to provide DRPs in the quantities needed for pharmacological (cell assays, animal studies), stability, and toxicity/safety studies in the early development stages.
Super Secrete
DRPs have been produced mainly by chemical synthesis (peptide synthesis). However, the chemical synthesis method makes it difficult to synthesize large amounts of DRPs with multiple disulfide bonds correctly formed in the molecule and requires high production costs. This has been a significant hurdle for DRP drug discovery in speeding up pharmacological evaluation and toxicity/safety assessment.
In collaboration with a Japanese chemical company, we have developed the Super Secrete method, a technology to express DRPs in the periplasmic space of E. coli and secrete them into the culture medium after folding them into the correct structure.
Generally, E. coli cannot secrete peptides or proteins into the culture medium, but the Super Secrete Method makes this possible using a unique culture medium. In purifying peptides and proteins by disrupting E. coli, a careful purification process is required to avoid contamination of E. coli-derived components such as LPS. Super Secrete, on the other hand, does not require disruption of E. coli, thus significantly reducing contamination of bacteria-derived components and simplifying the purification process. At the laboratory level, we have established a small-scale production method (culture and purification using a small jar fermenter).
Super Secrete enables us to provide DRPs in the quantities needed for pharmacological (cell assays, animal studies), stability, and toxicity/safety studies in the early development stages.
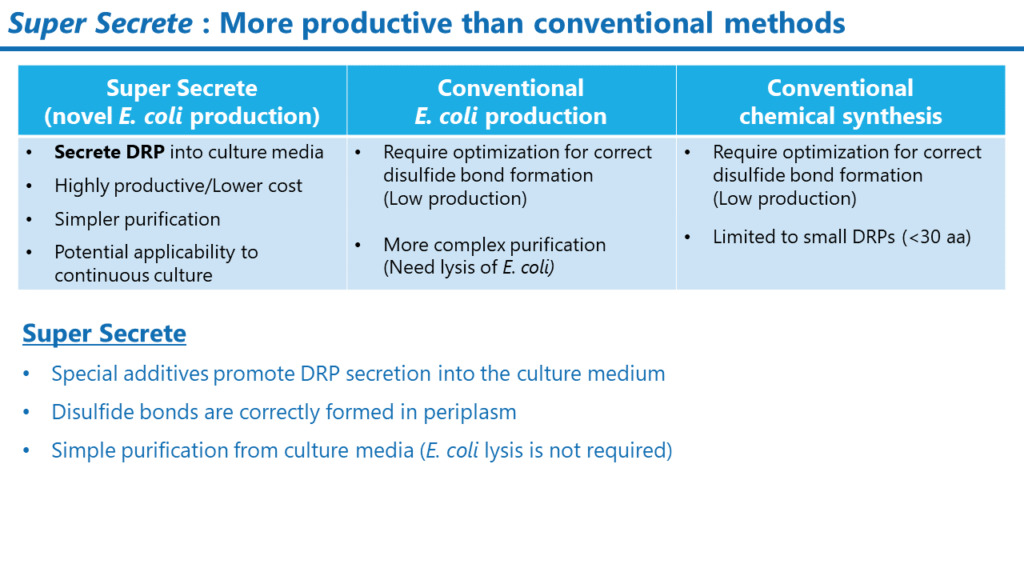
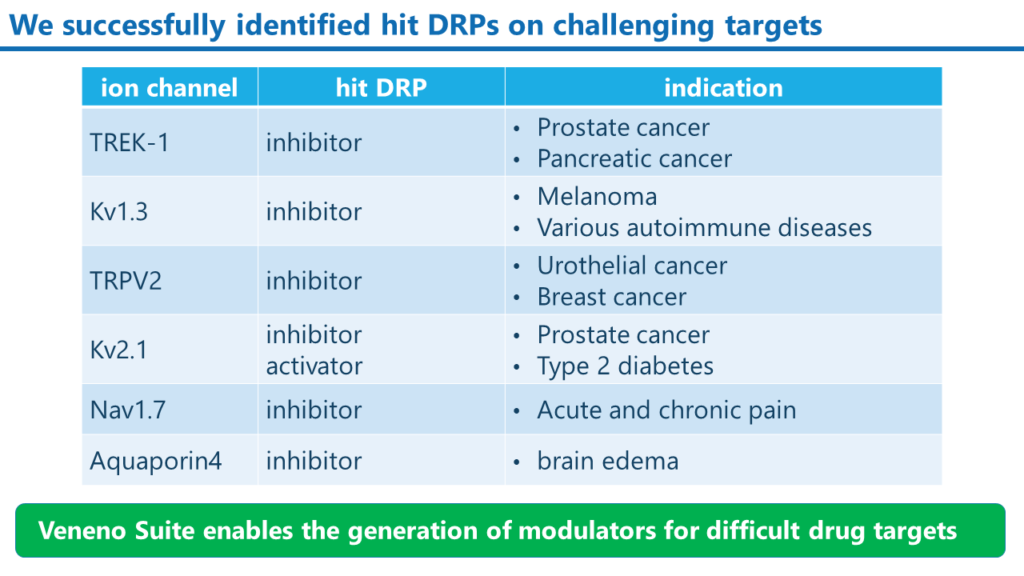
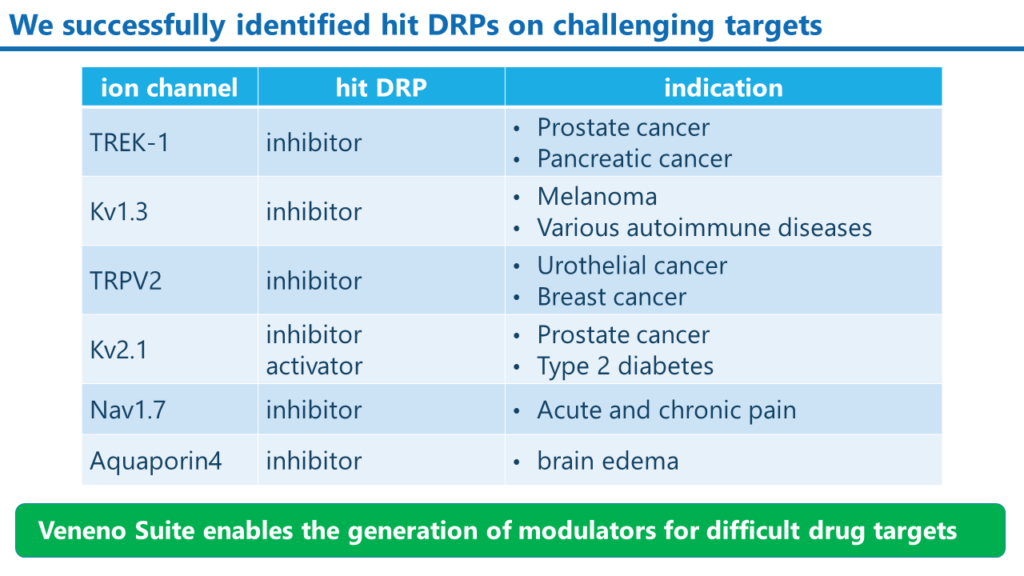
Examples of modulator DRPs that we have created
We have successfully created inhibitors and activators targeting various ion channels with our Veneno Suite.
Examples of modulator DRPs that we have created
We have successfully created inhibitors and activators targeting various ion channels with our Veneno Suite.


Veneno Technologies Co. Ltd.
2-1-6 Sengen Tsukuba, Ibaraki, 305-0047, Japan
info@veneno.jp